Background
SMILES (simplified molecular-input line-entry system) strings are representations of chemical structures often found in scientific literature. For example, water can be represented as [OH2] or [H]O[H] and glucose can be represented as OC[C@@H](O1)[C@@H](O)[C@H](O)[C@@H](O)[C@H](O)1.
Given the structured nature of these strings (they must contain valid chemical elements in valid positions) they make for an interesting use case for exploring the development of a LSTM-based binary classifier.
LSTMs are a form of recurrent neural networks that allow information to pass between training epochs, which makes them great for learning sequences. That’s great here because we want to consider SMILES strings as sequences, rather than simple bags of characters.
To support this project I’ve built a binary-class string classification framework, aptly named BinaryStringClassifier, which you can find on GitHub.
Framework
BinaryStringClassifier consists of two major components, each with their own sub-components:
BSC-Data: Entrypoint for dataset creation and curation.create: Create randomly generated datasets within given parameters.combine: Combine multiple components datasets into one.evaluate: Generate summary information for a given dataset.split: Perform train/test split for a given dataset.
BSC-Model: Entrypoint for model training & evaluation.train: Train a BinaryStringClassifier model.evaluate: Evaluate a trained BinaryStringClassifier model.predict: Get SMILES probabilities with a trained BinaryStringClassifier model
Process
Data Preparation
SMILES strings were manually pulled from various online sources, such as Wikipedia.
Negative class data (i.e. those that aren’t SMILES strings) are a combination of:
- English words
- Random strings
- Generated with
BSC-Data create -o ../data/random_strings.txt
- Generated with
- Random numbers
- Generated with
BSC-Data create -o ../data/random_digits.txt -c 1234567890 -l 1 10
- Generated with
- Manually curated false positives
- Identified during initial training
- e.g. words within brackets
Our combined dataset has the following metrics (generated with BSC-Data evaluate -d ./data/combined.csv):
| Metric | Count |
|---|---|
| Total entries | 404 |
| Non-SMILES | 327 |
| SMILES | 77 |
Note that we have a notable class imbalance here, with ~4.2 times more non-SMILES than SMILES strings. Our total dataset is also fairly small (n=404). Whilst generating new negative cases is pretty straight-forward, it’s trickier to collect positive examples of SMILES strings.
Building the model
There’s nothing too crazy about the architecture of our LSTM model:
model = Sequential()
model.add(Embedding(
input_dim=self.max_num_inputs,
output_dim=256,
input_length=self.max_input_size
))
model.add(SpatialDropout1D(0.3))
model.add(LSTM(256, dropout=0.3, recurrent_dropout=0.3))
model.add(Dense(256, activation='relu'))
model.add(Dropout(0.3))
model.add(Dense(2, activation='softmax'))
model.compile(
loss='categorical_crossentropy',
optimizer='Adam',
metrics=['accuracy']
)
Training the model
And training is as easy as:
BSC-Model train ./data/combined_train.csv -d trained_model -s 1337
Best model (so far)
After a few initial expansions of our training data to include some striking false negatives (mostly words in brackets would incorrectly classify as SMILES), we finally settle on a decent model that we can evaluate:
BSC-Model evaluate ./trained_model -d ./data/combined_test.csv
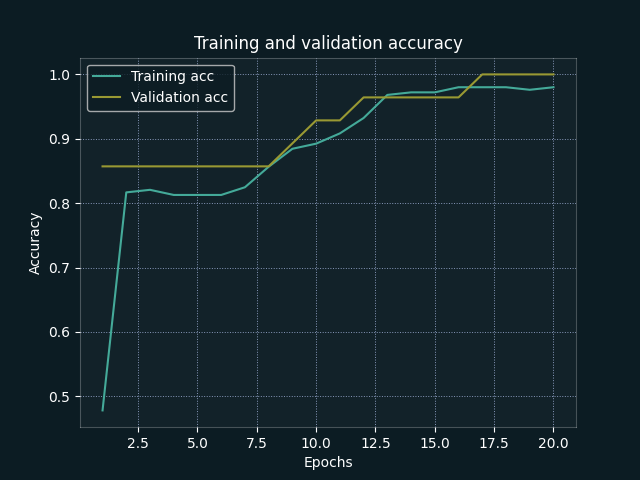
| Accuracy by Epoch | Loss by Epoch |
|---|---|
 |
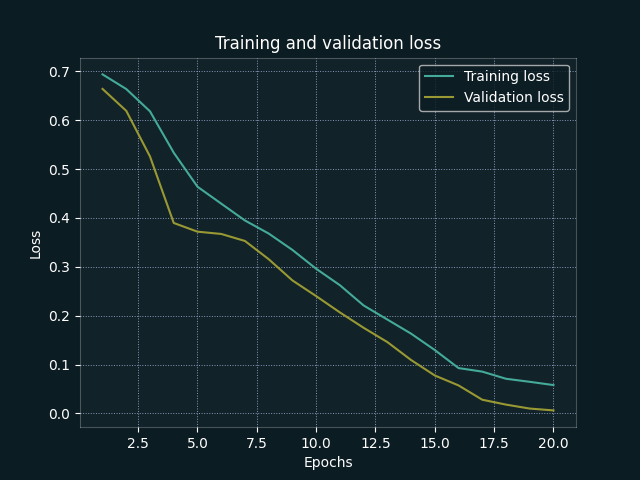 |
From our training evaluations, we can see that both training and validation accuracy increase together (suggesting that we are not overfitting to the training data). We also achieve strong metrics at epoch 20 (this epoch value was tweaked during initial training sessions), but a more generalised framework may benefit from implementation of early stopping.
| Metric | Value |
|---|---|
| Accuracy | 0.992 |
| Precision | 1.000 |
| Recall | 0.960 |
| F Score | 0.980 |
| Final Validation Accuracy | 1.000 |
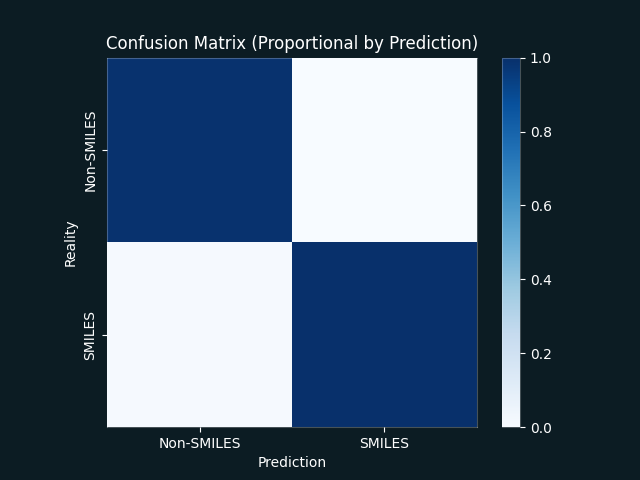
Only one misclassification was identified in our test dataset (C#N was not recognised as a valid SMILES).
| Confusion Matrix | ROC Curve |
|---|---|
 |
 |
Ultimately we’re looking at an insanely good set of metrics that either suggest our model has identified a strong signal that differentiates SMILES and non-SMILES strings (and therefore that our LSTM may be overkill) or (possibly more likely) that we don’t really have enough data to train and test on and will fail to generalise on a larger real world dataset.
Either way, we now have a framework through which additional data can be trained on and that can be modified for similar classification tasks.
Full Run Script
# Data Preparation
## Create dataset components (not seeded)
BSC-Data create -o ../data/random_strings.txt
BSC-Data create -o ../data/random_digits.txt -c 1234567890 -l 1 10
## Combine datasets
BSC-Data combine \
-f ./data/random_strings.txt \
./data/random_words.txt \
./data/random_digits.txt \
./data/refine_FP.txt \
-s ./data/smiles.txt \
-o ./data/combined.csv
## Validate final dataset & generate summary stats
BSC-Data evaluate -d ./data/combined.csv
## Create train/test splits
BSC-Data split \
-d ./data/combined.csv \
-t 0.3 \
-s 1337
# Model Training
## Train model
BSC-Model train ./data/combined_train.csv -d trained_model -s 1337
## Evaluate model
BSC-Model evaluate ./trained_model -d ./data/combined_test.csv
## Run predictions
BSC-Model predict ./trained_model -s strings_for_prediction.txt