Background
Malawi experienced prolonged use of sulfadoxine/pyrimethamine (SP) as the front-line anti-malarial drug, with early replacement of chloroquine and delayed introduction of artemisinin-based combination therapy. Extended use of SP, and its continued application in pregnancy is impacting the genomic variation of the Plasmodium falciparum population.
Methods
Whole genome sequence data of P. falciparum isolates covering 2 years of transmission within Malawi, alongside global datasets, were used. More than 745,000 SNPs were identified, and differences in allele frequencies between countries assessed, as well as genetic regions under positive selection determined.
Results
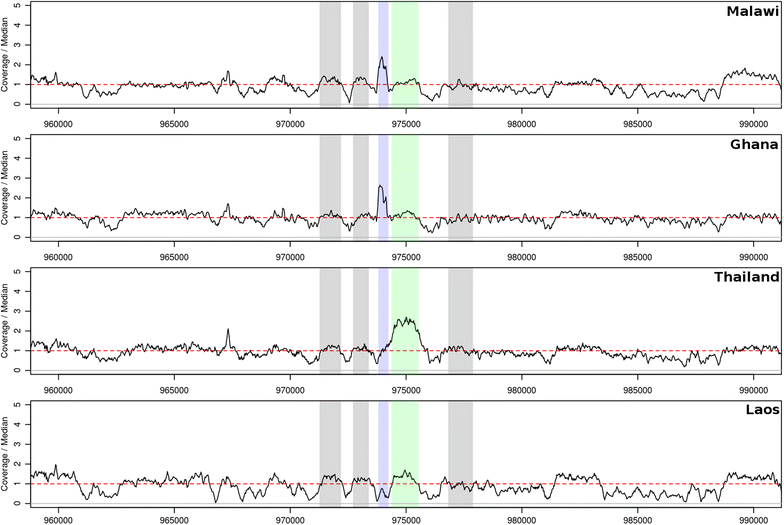
Positive selection signals were identified within dhps, dhfr and gch1, all components of the parasite folate pathway associated with SP resistance. Sitting predominantly on a dhfr triple mutation background, a novel copy number increase of ~twofold was identified in the gch1 promoter. This copy number was almost fixed (96.8% frequency) in Malawi samples, but found at less than 45% frequency in other African populations, and distinct from a whole gene duplication previously reported in Southeast Asian parasites.
Conclusions
SP resistance selection pressures have been retained in the Malawian population, with known resistance dhfr mutations at fixation, complemented by a novel gch1 promoter duplication. The effects of the duplication on the fitness costs of SP variants and resistance need to be elucidated.
Full Text
Citation: Matt Ravenhall, Ernest Diez Benavente, Mwapatsa Mipando, Anja T. R. Jensen, Colin J. Sutherland, Cally Roper, Nuno Sepúlveda, Dominic P. Kwiatkowski, Jacqui Montgomery, Kamija S. Phiri, Anja Terlouw, Alister Craig, Susana Campino, Harold Ocholla and Taane G. Clark. Characterizing the impact of sustained sulfadoxine/pyrimethamine use upon the Plasmodium falciparum population in Malawi. Malaria Journal 201615:575